File format used by the dmfit program
Reading
The "dmfit" & "Edit NMR" programs can read in
different file formats that they read in using automatic detection routines :

Bruker Formats [1r 2rr 3rrr fid ser and .1r .rr .rrr .fid .ser]
-
Native Bruker XWinNMR format from the spectrometer (Unix, WindowsNT and Linux ) (read only)
-
WinNMR (Windows PC) file format (read and save)
more details on the Bruker Formats...

ASCII files [.txt .hz .ppm .asc]
-
Ascii files is a list of X and Y coordinate with even spacing in X - Hz (read and save)
download an example
typical file :
ti: string title
##freq <frequency in MHz> // optional
x1 y1
x2 y2
...
xn yn
Important remark :
+ the dX increment is computed from xn and x0 dX = (xn-x0)/(n-1)
+ the sweep width Sw is taken as n*dX to be compatible with Fourier transfromed
data

SIMPSON like 1D and 2D [.spe]
File extension is ".spe".
dmfit can read SIMPSON ascii coded data (no binary coding).
It uses a SIMPSON like format to read/save computed intermediate spectra (stored in the temp directory of the computer)
It can read/save in this format.
more information and examples for Simpson like Formats...
Core Scientific Data Format - CSDF - under development
The CSDF format is a Json based format that we (Philipp Grandinetti - RMN, Thomas Vosegaard - SIMPSON and related tools, Dominique Massiot - dmfit) currently develop to allow cross exchange of data between processing, fitting or modeling softwares...
more details on the Csdf Formats...
Saving
The "dmfit" & "Edit NMR" programs only save using
the following formats :
+ WinNMR (Windows PC) file format
+ Ascii files (list of X and Y coordinate
with even spacing in X - Hz)
Native Bruker XWinNMR format
|
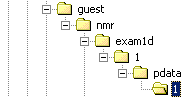
guest is the username
exam1d is the experiment name
1 is the experiment nb, it contains fid, aqus, acqu2s, ser,
pulprogram files
pdata contains the processed data
1 is the process nb, it contains the 1r, 1i, procs, 2rr, 2ii, 2ir, 2ri,
proc2s files and many others
|
WinNMR PC format
All files reside in a single directory named with the experiment
name.
eeeppp.xxx correspond to experiment e and process p files (007001.xxx is exp7
proc 1)
dmfit and EditNMR programs accept names as well as numbers
The correspondance between XWinNMR is the following :
| 1D dataset |
|
|
| XWinNMR |
WinNMR PC |
Content |
| acqus |
007001.aqs |
acquisition paramters |
| procs |
007001.fqs |
process parameters |
| fid |
007001.fid |
raw fid data points |
| 1r |
007001.1r |
spectrum real part data points |
| 1i |
007001.1i |
spectrum imaginary part data points |
| 2D dataset |
|
|
| XWinNMR |
WinNMR PC |
Content |
| acqus |
007001.fa2 |
acquisition paramters F2 |
| procs |
007001.fp2 |
process parameters F2 |
| acqu2s |
007001.fp1 |
acquisition paramters F1 |
| proc2s |
007001.fp2 |
process parameters F1 |
| ser |
007001.ser |
raw ser data points |
| 2rr |
007001.rr |
2D spectrum real part data points |
| 2ii |
007001.ii |
2D spectrum imaginary part data points |
The acqu & proc files are now in ascii and were previously
binary coded, dmfit and EditNMR can read and save both.
fid, ser, 1r, 1i, 2rr, 2ii, 2ri and 2ir contain integers with encoding depending
upon the OS used.
WinNMR data files (ser, fid, 1r, 1i, rr, ii...) are usually stored in float
format.