How to Fit an MQMAS spectrum

Example :
87Rb MQMAS spectrum of RbNO3
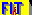

Example :
27Al Syncrhonized MQMAS
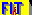

Example :
27Al 2D MQMAS Spectrum of Glass with AlO4/AlO5/AlO6 sites
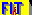

Example :
27Al STMAS Spectrum of A9B2 at 830MHz
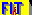
Quadrupolar tensor definition :
-
principal values are sorted |Vyy|<=|Vxx|<=|Vzz| note that Vxx and Vyy have
signs opposite to that of Vzz - usually NMR is not sign sensitive for EFG
-
Vzz+Vxx+Vyy=0
-
etaQ is the anisotropy of the Quadrupolar tensor etaQ=Vyy-Vxx/Vzz with
0<=EtaQ<=1
-
and thus Vxx=1/2 (-EtaQ-1)Vzz and Vyy=1/2 (EtaQ-1)Vzz

|
Under construction - Construction en cours....
Old link - Ancien lien :
Remarks & Comments - Remarques & commentaires : email
|
Step 1 : Calibrate the MQ-MAS experiment
The fit of the MQ-MAS
(or STMAS) experiment assumes that you sheared the acquired spectrum (with
Bruker AU program, with RMN Grandinetti's program, or any other way...).
The F1 axis is
assumed to be scaled similarly with the F2 axis so that the diagonal (slope 1)
1/1 line relates identical isotropic chemical shifts.
For datasets
acquired with Bruker, using "in0" as the increment you can use the calibration
algorithm :
-
Call the
SpecParam dialog [Menu/File/View Param]
-
Set the proper
nucleus (??? list box second line) which has to be quadrupolar !!
-
Click the
"Calibrate MQ..." button at the bottom of the dialog.
-
Accept the
values proposed in the "Calibrate MQ experiment" dialog (in which you can
specify the shift that you applied or the folding)
Remark
: depending on your pulse sequence and on the considered nucleus, it can happen
that the slope of the chemical shift diagonal is negative, in that case you have
to reverse the F1 dimension : [Menu/2D/Reverse F1]
Step 2 : if possible begin by Fitting the 1D spectrum.
This step is
not necessary but can simplify the process of fitting the 2D dataset.
If you need
you can learn more about fitting a 1D
Quadrupolar broadened spectrum.
Step 3 : Switch to the 2D dataset.
Having the 1D
spectrum and fit loaded, just go the the [Menu/File/Open] and open the 2D
dataset (*.rr files for Bruker).
or
Load in the 2D dataset [Menu/File/Open] and open the 2D dataset (*.rr files for
Bruker) and load the 1D fit parameters [Menu/Decomposition/Load Other]
Step 4 : Switch the decomposition mode from 1D to
2D
In the lef
panel, change the fitting mode from "Fit 1D" to "Fit MQ" (if you do not see this
option, enlarge the dmfit window to make it appear).
Click the
 and you should get the first guess
and you should get the first guess
Step 5 :